Skip to:
- How does SMRT Sequencing Work?
- Studying DNA Methylation in Bacteria; an Application of SMRT Sequencing
DNA sequencing works by using DNA polymerase to add nucleotides to a template. There are several technologies available for DNA sequencing. One such example is Single-Molecule Real-Time sequencing, or SMRT sequencing.
How does SMRT Sequencing Work?
As with other DNA sequencing technologies, the first step after DNA extraction is to prepare a “library”. This process prepares the DNA for the sequencing; in this case, adaptors are added to either end of a double stranded DNA molecule, which effectively enables the DNA to become a single stranded circular template. This then means that the DNA can be sequenced continuously.
This DNA library, or the template DNA, is then put into a DNA sequencer which contains “zero-mode waveguides” which have DNA polymerase immobilized at one end. A single DNA molecule is then immobilized into these zero-mode waveguides, and the DNA polymerase starts adding new nucleotides to a de novo synthesized DNA strand complementary to the template DNA. The bases in these nucleotides are labelled, and incorporation of these bases into the growing DNA strand causes light emission. This light emission is then read in real-time, and as the emission from each base is different this allows for the specific base to be identified.
The main advantage of SMRT sequencing is the generation of long sequencing reads of high accuracy, which improves the assembly of whole genomes. This is because longer sequencing reads mean less “building” is required to assemble the genome.
Studying DNA Methylation in Bacteria; an Application of SMRT Sequencing
What is DNA methylation?
The addition of a methyl group to DNA, also known as methylation, occurs in all kingdoms of life. There are three methylated nucleotides present in bacteria; m5C (C5-methyl-cytosine, which is also present in eukaryotes), m6A (N6-methyl-adenine) and m4C (N4-methyl-cytosine, which is only found in bacteria). Methylation occurs after the synthesis of new DNA strands, and happens at specific nucleotides.
The methyl groups stick out of the DNA double helix, and therefore can impact the binding between DNA and DNA binding proteins. This in turn impacts processes including chromosome replication, DNA mismatch repair, as well as timing of gene transcription and formation of epigenetic lineages.
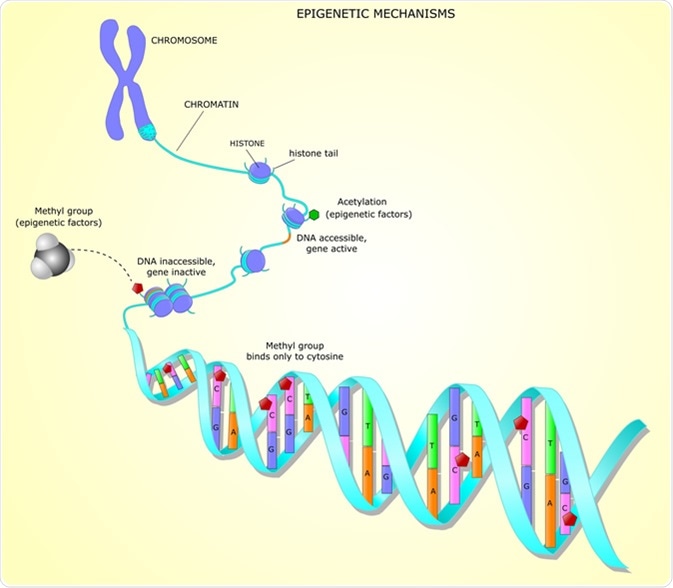
Why is DNA methylation important in bacteria?
Bacteria are infected by viruses, therefore need a protective mechanism to overcome viral infections. This is where restriction-modification systems come into play; this system is made up of a restriction enzyme, which breaks down DNA at specific sites, and a DNA methyltransferase, which adds a methyl group to adenine (A) or cytosine (C).
In the majority of restriction-modification systems, the DNA methyltransferase acts to protect the bacterial DNA from the restriction enzyme. The presence of the DNA methyltransferase means that the bacterial DNA becomes methylated, while the infecting viral DNA is not. This in turn means that the viral DNA is degraded by the restriction enzyme, while the bacterial DNA is protected due to the restriction enzyme not acting on methylated DNA. However, it should be noted that there are restriction enzymes which act on modified DNA.
Recent studies have hinted that there may be additional roles for restriction-modification systems. For example, knocking out certain restriction-modification systems resulted in a change in gene expression, which is linked to the difference in DNA methylation. Restriction-modification systems can also cause double-stranded breaks and C-T mutations, thereby influencing the evolution of bacteria. More recently, technologies have been developed that are able to determine the methylation of an entire bacterial genome, known as the “methylome”.
How do you determine the methylome using SMRT sequencing?
As SMRT sequencing gives real-time results, it can be used to detect DNA modifications including methylations. The DNA polymerase incorporates nucleotides at a constant rate, but this rate can be changed if the nucleotide in the template had been modified. This can be noted during the sequencing process.
Blow at al. used SMRT sequencing to map DNA modifications in 230 microorganisms. The modifications they looked for included m5C, m6A and m4C. The authors found that 93% of these microorganisms showed DNA methylation, and they also found 834 motifs which were methylated. This enabled the authors to identify which motifs are the targets for 620 DNA methyltransferases.
Interestingly, the authors noted that while 48% of the studied organisms had a DNA methyltransferase, there was no evidence of a restriction enzyme also being present. Therefore, it is possible that DNA methylation plays an important role in genome regulation, or another important role in microorganisms which is yet to be identified.
Sources
- PacBio. SMRT sequencing brochure www.pacb.com/…/…-long-reads-to-drive-discovery-in-life-science.pdf
- PacBio. SMRT sequencing – how it works www.pacb.com/…/Infographic_SMRT-Sequencing-How-it-Works.pdf
- Sánches-Romero, M. A. et al., DNA methylation in bacteria: from the methyl group to the methylome. Current Opinion in Microbiology 2015, 25, 9-16. https://www.sciencedirect.com/science/article/pii/S1369527415000399
- PaBio. SMRT Sequencing: Epigenetics https://www.pacb.com/smrt-science/smrt-sequencing/epigenetics/
- Blow, M. J. et al. The Epigenomic Landscape of Prokaryotes. PLOS Genetics 2016, 12 (2), e1005854. journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1005854
Further Reading
- All DNA Sequencing Content
- DNA Sequencing
- DNA Sequence Assembly
- DNA microarray
- High-throughput DNA Sequencing Techniques
Last Updated: Sep 3, 2019

Written by
Dr. Maho Yokoyama
Dr. Maho Yokoyama is a researcher and science writer. She was awarded her Ph.D. from the University of Bath, UK, following a thesis in the field of Microbiology, where she applied functional genomics toStaphylococcus aureus . During her doctoral studies, Maho collaborated with other academics on several papers and even published some of her own work in peer-reviewed scientific journals. She also presented her work at academic conferences around the world.
Source: Read Full Article