 Thought LeadersDr David SünderhaufPostdoctoral Research FellowUniversity of Exeter
Thought LeadersDr David SünderhaufPostdoctoral Research FellowUniversity of ExeterIn this interview, we spoke to Dr. David Sünderhauf, a microbiologist and a postdoctoral research fellow at the University of Exeter, about his latest research removing AMR plasmids using a mobile, broad-host-range CRISPR-Cas9 delivery tool.
Please introduce yourself, tell us about your career background, and provide insight into what has inspired your career to date?
Hiya! My name is David Sünderhauf (prev Walker-Sünderhauf), I’m a microbiologist and a postdoctoral research fellow at the University of Exeter. During my postdoc and before that my PhD, I’ve been researching novel approaches to tackle antimicrobial resistance. I’m particularly interested in antibiotic-resistance genes carried on mobile genetic elements. Apart from just this research (as exciting as it is!), I’m also inspired by outreach and science communication.
Posing a threat to human and animal health, the environment, food security, economic development, and societal equity, antimicrobial resistance (AMR) has been listed as one of the top 10 threats to health. Please could you outline what AMR is and why it poses such a significant global threat?
AMR is an umbrella term that describes when microbes, mostly bacteria or fungi, are resistant to the drugs we use to treat them. This means, thanks to AMR, antibiotics that we have been routinely using since the 1940s often don’t work anymore. For now, antibiotics can still often be used, but this is quickly changing with many multi-drug resistant infections seen every day. Without effective antibiotics, we’d be back in a world where a simple cut can mean that you need to lose your arm, or where complex surgeries are completely impossible.
AMR is such a complex global issue because it is the bacteria (rather than people) that are becoming resistant to antibiotics – and bacteria can exchange genetic material that makes them resistant, giving rise to “superbugs” that can’t be treated with any type of antibiotic. This means mis- and overuse of antibiotics in one part of the environment or one part of the world can have global impacts.
There currently is a global effort across many different sectors to tackle the spread and lessen the impact of AMR. What role does research play in this fight, and why is research so important?
As I said above, AMR is complex and can’t just be summarized as an issue for the clinic, which is why it is great that there is an effort across different sectors to tackle this problem.
There are many different types of research into AMR going on that in my opinion are all important to solve the problem: there is no one miracle solution that will solve everything on its own. Some of these research avenues are, for example, research into new antibiotics, research into how to most effectively use existing antibiotics and stop resistance arising, and finally research into alternatives to antibiotics.
Without research into these three different areas, any other fix (for example providing better diagnostics to make sure antibiotics are only used when necessary) would be just temporary. Bacteria still evolve, and antibiotic resistance genes still spread, meaning superbugs will still cause problems.
 Image Credit: angellodeco/Shutterstock.com
Image Credit: angellodeco/Shutterstock.com
By 2050, AMR is estimated to cause more human deaths than cancer and diabetes combined. High-quality research is necessary to develop an effective response to the issue. Your most recent study involves a well-known gene editing technique known as CRISPR-Cas 9. Could you please outline what CRISPR-Cas 9 is and how it works in the context of eliminating AMR?
In our work, we use CRISPR-Cas9 to remove antibiotic resistance genes. In nature, CRISPR-Cas9 is a bacterial immune system that can learn to recognize specific DNA sequences and cut these. When using it as a biotechnology, we can repurpose this by “telling” Cas9 which DNA sequence to recognize and to cut. We programmed Cas9 to target an antibiotic resistance gene, which means bacteria that have received this CRISPR-Cas9 treatment will lose the antibiotic resistance gene that is being targeted and can be treated by antibiotics again.
Your most recent study utilizes a broad-host-range CRISPR-Cas9 delivery tool to remove AMR plasmids from multiple members of complex microbial communities. Please could you outline how you conducted this research and tell us the main outcomes of this study?
Plasmids are short pieces of DNA that bacteria can often exchange with each other – this is one of the main avenues which antibiotic-resistance genes can get passed on between different bacterial species. We engineered one such plasmid to encode CRISPR-Cas9. The plasmid we chose, pKJK5, is known to have a very broad host range which means it can be taken up by many different species of bacteria, including pathogens such as Escherichia coli, Pseudomonas aeruginosa, or Staphylococcus aureus.
In this study, we carry out proof-of-concept experiments and show that we can use our CRISPR-Cas9 tool to protect a series of different isolates from the uptake of an antibiotic resistance plasmid, which prevents them from becoming resistant. We also show that we can remove an antibiotic resistance gene that is already present in a target E. coli bacterium – this means we can use this CRISPR-Cas9 tool to resensitise bacteria that were previously resistant to antibiotics, and successfully kill them with this antibiotic after the CRISPR-Cas9 treatment.
The exciting part is firstly the broad host range of this CRISPR-Cas9 tool, which means we can apply it in a range of different bacteria without having to re-engineer it. Secondly, the way it can be delivered to target bacteria is very ecologically feasible. Because we engineered a natural plasmid, which bacteria naturally exchange with each other, this CRISPR-Cas9 tool can be delivered to microbiomes by adding a “donor strain”, essentially a probiotic bacterium that already carries the CRISPR-Cas9 tool and will pass it on to other bacteria.
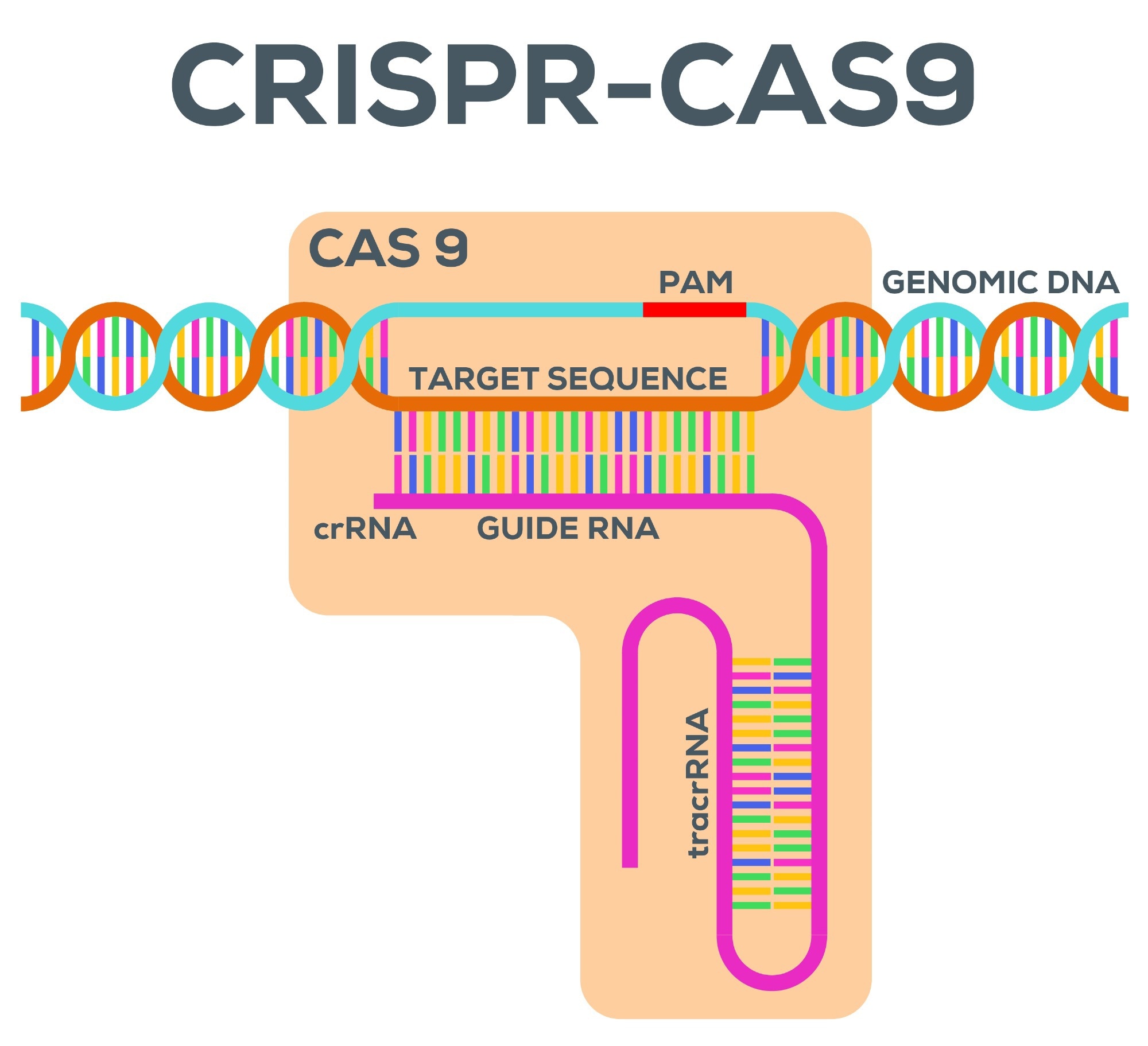 Image Credit: Freakeistein/Shutterstock.com
Image Credit: Freakeistein/Shutterstock.com
AMR is a complex, continually evolving issue that disproportionately affects low- or middle-income countries (LMICs). Affordable and accessible techniques to combat AMR are required to bridge the uneven societal burden of AMR. While CRISPR-Cas-9 provides a novel approach to tackle the issue, do you believe CRISPR is an accessible approach that could be utilized in LMICs to slow the growing spread of multi-resistant bacteria? How can the potential for inequitable access be avoided?
Absolutely, once fully developed I don’t see any reason why a technology like this should stay inaccessible to LMICs – especially, as mentioned above, as AMR is a global issue that needs to be tackled globally. Once this technology is developed for more specific use cases, I think it will be very important to engage directly with the people who will be using it, in the communities that will be using it. And this of course includes LMICs.
Your most recent study establishes a promising broad host-range CRISPR-Cas9 delivery tool for AMR plasmid removal, which has the potential to be applied in complex microbial communities to remove AMR genes from a broad range of bacterial species. What could this mean for the future of AMR?
A tool like this has the potential to be used in two different main applications. In clinics, it could be used to make sure a patient’s infection is not resistant to antibiotics so that treatment afterward will be successful. In the environment, it could be used in places like waste-water treatment plants, where bacteria often exchange genetic material with each other, including antibiotic resistance genes. Using a CRISPR-Cas9 tool like ours, we could interfere with this process and stop superbugs arising in the first place, before they cause problems in the hospital.
What further steps are required for this tool to be applied in real-world settings to eliminate AMR genes from bacterial communities?
There are several technical challenges we are still facing, one of these is the delivery efficiency – how well does the CRISPR-Cas9 tool transfer into the bacteria in which we want it to act?
Apart from this sort of research, legislation, and policymakers need to be engaged. CRISPR-Cas9 is a bacterial immune system, which we use to “immunize” bacteria from antibiotic resistance genes.
Nevertheless, we are still using an engineered plasmid to deliver this to the target bacteria. Application of a technology like this should of course only be carried out in line with regulations (which need to be made), and after stakeholder engagement of people it directly affects.
What's next for you and your research? Are you involved in any exciting upcoming projects?
We’re still very actively working on this CRISPR-Cas9 delivery tool and applying it in the lab in more complex communities, against more clinically relevant antibiotic resistance genes, and figuring out different approaches to getting around target bacteria properties that might limit how effective the treatment is.
Apart from this, we are also discovering some cool things about the fundamentals of how plasmids (the accessory pieces of DNA that carry antibiotic-resistance genes) interact with each other, and how plasmids are maintained and spread through bacterial communities. This sort of insight adds to a wealth of other work and can allow us to predict how antibiotic-resistance genes might spread in different microbiomes.
Where can readers find more information?
- https://www.microbiologyresearch.org/content/journal/micro/10.1099/mic.0.001334
- https://vanhoutelab.wordpress.com/
- https://twitter.com/davvi36
About Dr. David Sünderhauf
David is a Postdoctoral Research Fellow at the University of Exeter, UK. He holds a PhD as well as an MRes in Microbiology from the University of Exeter, and a BSc (Hons) in Immunology from the University of Aberdeen.
Apart from his research, he is also involved in organizing public science events as co-chair of Cornwall Science Community, and in local politics as a Green Party Truro City Councillor.
Posted in: Thought Leaders
Tags: Antibiotic, Antibiotic Resistance, Antimicrobial Resistance, Bacteria, Biotechnology, Cancer, Cas9, CRISPR, Diabetes, Diagnostics, DNA, Drugs, E. coli, Food, fungi, Gene, Genes, Genetic, Hospital, Immune System, Immunology, micro, Microbiology, Plasmid, Probiotic, Research, Science Communication, Staphylococcus aureus, Technology
Source: Read Full Article