With a rise in antibiotic resistance towards the existing range of antibiotics, there is a need to discover a new class of previously unknown antibiotic peptides.
Previous methods of identifying new antibiotics involve procedures that are slow, inefficient, and focuses on cationic (positively charged) and amphipathic peptides. Thus, there is a need to discover newer methods to uncover diverse range of antibiotic peptides.
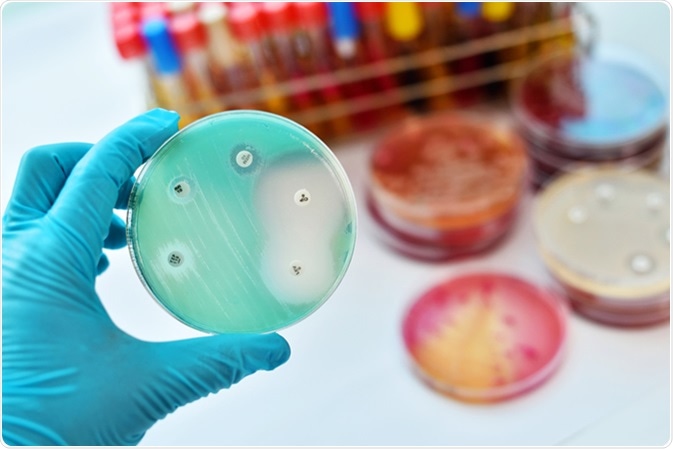
Surface localized antimicrobial display or SLAY involves construction of a peptide library of random peptide sequences, which are then transformed or incorporated in to a bacterial genome (E. coli).
Then these peptide sequences are induced by IPTG and screened for antibiotic effects, as the bacteria which expresses antibiotic peptides will be eliminated from the population.
Subsequently using next generation sequencing, the sequences before and after the induction are derived and analysed in silico to determine the top antibiotic peptides.
As a drug first interacts with the bacterial cell surface, all the peptides transformed during SLAY have a surface localisation signal.
High-Throughput Technique to Detect Large Number of Antibiotics
The main potential of SLAY technique lies in its high-throughput nature. Firstly, peptides of any size, structure, or sequence complexity are driven in the gram-negative bacteria. These peptides are then expressed and the result is read using high-throughput DNA sequencing.
This helps in screening large number of potential peptides in a small amount of time. Researchers at University of Texas used SLAY to screen a massive and unbiased peptide library of random sequences and probed for antimicrobial activity.
They were able to quickly screen approximately 800,000 20-mer peptides for antimicrobial activity. Although a vast majority (98.3%) of sequences did not show any antibiotic effects, 1.7% (7,968 fully synthetic sequences) of the pool did show potential antimicrobial activity.
This single screen doubled the number of known antibiotics discovered till now. This scale of antibiotic peptide search is almost impossible using traditional methods of antibiotic screening.
Discovery of Chemically Diverse Antibiotics
Due to lack of simple and biologically relevant methods to screen for antibiotic, most of the current strategies focus on a single class of cationic (positively charged) and amphipathic (containing both hydrophilic and hydrophobic regions) peptides or CAMPs (cationic antimicrobial peptides).
However, when SLAY peptides were tested for their charge and hydrophobicity, there was no bias towards positively or negative charge in these compounds.
Most of the compounds had neutral charge and neutral hydrophobicity. Also, the current peptides are dominated by lysine-rich sequences but there was no such specific enrichment of positively charged residues.
Hydrophobic residues, such as alanine, isoleucine, leucine, and valine were also equally present. Thus, SLAY uncovers new and untapped range of antibiotics which was previously dominated by a single class of peptides.
Discovery of Less Toxic Antibiotics
The existing peptides can be engineered to exhibit salt tolerance and resistance to proteases (enzyme that degrades proteins). However, it has been difficult to make existing CAMPs or cationic antimicrobial peptides less toxic.
One of the cationic peptides P1 shows seizure-like activity at 25 mg/kg and causes immediate mortality at 35 mg/kg. Thus, cationic peptides frequently show toxic effects.
Hemolysis (break down of RBCs), one of the read-outs of the toxicity shown by antibiotics, was tested in SLAY peptides and none of the peptides discovered through SLAY exhibited hemolytic or toxic effects up to a maximum dose of 50mg.
The Synthetic Form Shows Good Performance
Many of the antibiotic peptides discovered using the traditional approaches fail to show activity in synthetic form as they cannot cross the bacterial membrane. Thus, the performance of the SLAY peptide was tested to assess if they possess similar drawbacks.
To test this, SLAY peptides with low hydrophobicity and neutral to negative charge were selected. This was done in order to test if SLAY could show similar level of activity as the usually employed positively charged antibiotic peptides.
These were tested against three bacteria-resistant strains. 18 of 22 (80%) sequences which were identified by SLAY as active exhibited antimicrobial activity.
Atypical, non-cationic, non-hydrophobic peptides showed a range of antibacterial effects on the four bacteria tested, uncovering a new range of potential antibiotics.
Certain peptides also showed antimicrobial action towards some strains and no activity towards others. This suggests a more specific mode of action of SLAY peptides.
Sources:
- Discovery of Next-Generation Antimicrobials through Bacterial Self-Screening of Surface Displayed Peptide Libraries. Cell 2018 Jan 25;172(3):618-628.
- Antimicrobial Peptides: An Emerging Category of Therapeutic Agents.
Further Reading
- All Antibiotic Resistance Content
- Global travel and antibiotic resistance
- Horizontal Gene Transfer and Antibiotic Resistance
- Antibiotic Resistance Influence on Wound Care
- Unapproved Antibiotics and Antibiotic Resistance
Last Updated: Aug 23, 2018

Written by
Hannah Simmons
Hannah is a medical and life sciences writer with a Master of Science (M.Sc.) degree from Lancaster University, UK. Before becoming a writer, Hannah's research focussed on the discovery of biomarkers for Alzheimer's and Parkinson's disease. She also worked to further elucidate the biological pathways involved in these diseases. Outside of her work, Hannah enjoys swimming, taking her dog for a walk and travelling the world.
Source: Read Full Article