Researchers in the UK and Germany have provided evidence for a novel mechanism by which severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the agent that causes coronavirus disease 2019 (COVID-19) – binds to human host cells.
The team – from the Wellcome Sanger Institute in Cambridge, the University of Cambridge, and the Leibniz Institute for Primate Research in Göttingen – found that in addition to the host cell receptor angiotensin-converting enzyme 2 (ACE2), the spike protein of SARS-CoV-2 also binds to a protein encoded by the LRRC15 (leucine-rich repeat containing 15) gene.
Gavin Wright and colleagues say this interaction is distinct from previously known spike attachment factors and appears to have emerged recently within the coronavirus family.
Assays showed that while the expression of LRRC15 alone was not sufficient to permit viral entry, its interaction with the spike could modulate infection.
“Our data point to an unappreciated host factor for SARS-CoV-2, with potential relevance to COVID-19,” says the team.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.
.jpg)
More about the SARS-CoV-2 infection process
The binding interactions that occur between SARS-CoV-2 and host cell factors enable the virus to propagate infection that causes COVID-19. The unprecedented virulence and spread of SARS-CoV-2 suggest that the virus has developed particularly effective mechanisms for targeting host cells.
The most prominent structure on the surface of SARS-CoV-2 is the spike protein, which mediates the initial stage of infection when its receptor-binding domain (RBD) attaches to ACE2. This spike is a central component in many of the vaccines and antiviral therapies that have been developed to help prevent or treat COVID-19.
While the spike is already known to bind to ACE2, there is considerable debate regarding whether ACE2 alone is sufficient to explain COVID-19 pathology.
For example, certain target cells only express low levels of ACE2 and questions remain over whether additional factors may explain the broad cellular tropism of SARS-CoV-2.
What did the current study involve?
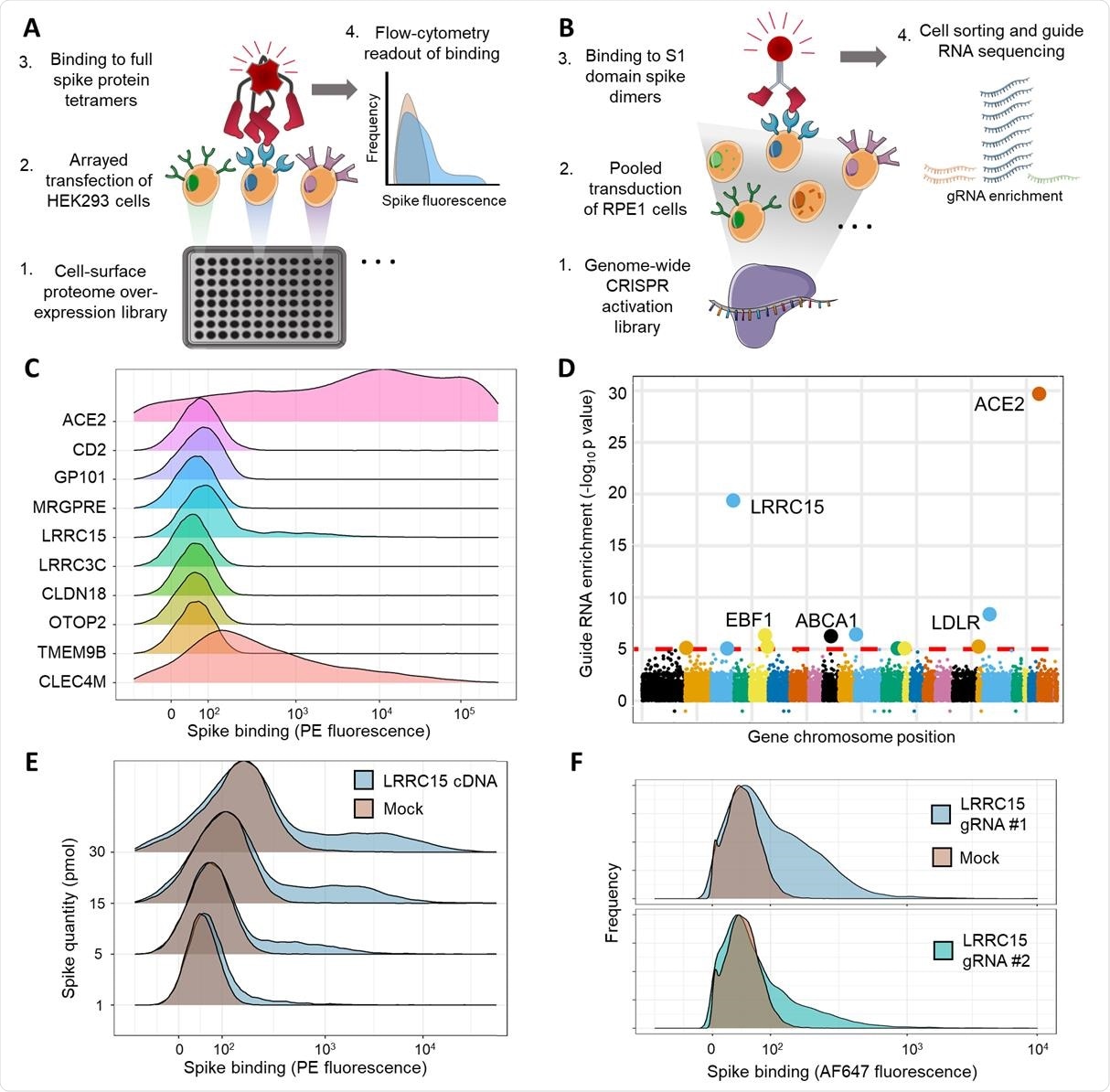
Wright and colleagues performed two independent cell-based screens of SARS-CoV-2 spike binding to determine whether host proteins other than ACE2 enable the spike to interact with cells.
This screening identified LRRC15 expression as a novel additional host factor that promotes spike binding.
The interaction of spike with LRRC15 was reproducible in different human cell lines and was distinct from other known glycan- or ACE2-binding pathways.
Analysis of related coronavirus spikes revealed that the interaction was restricted to SARS-CoV-2, suggesting that LRRC15 represents a novel class of spike binding interaction.
The interaction was located to the C-terminus of the S1 domain of the spike protein and LRRC15 shared recognition of the ACE2 RBD.
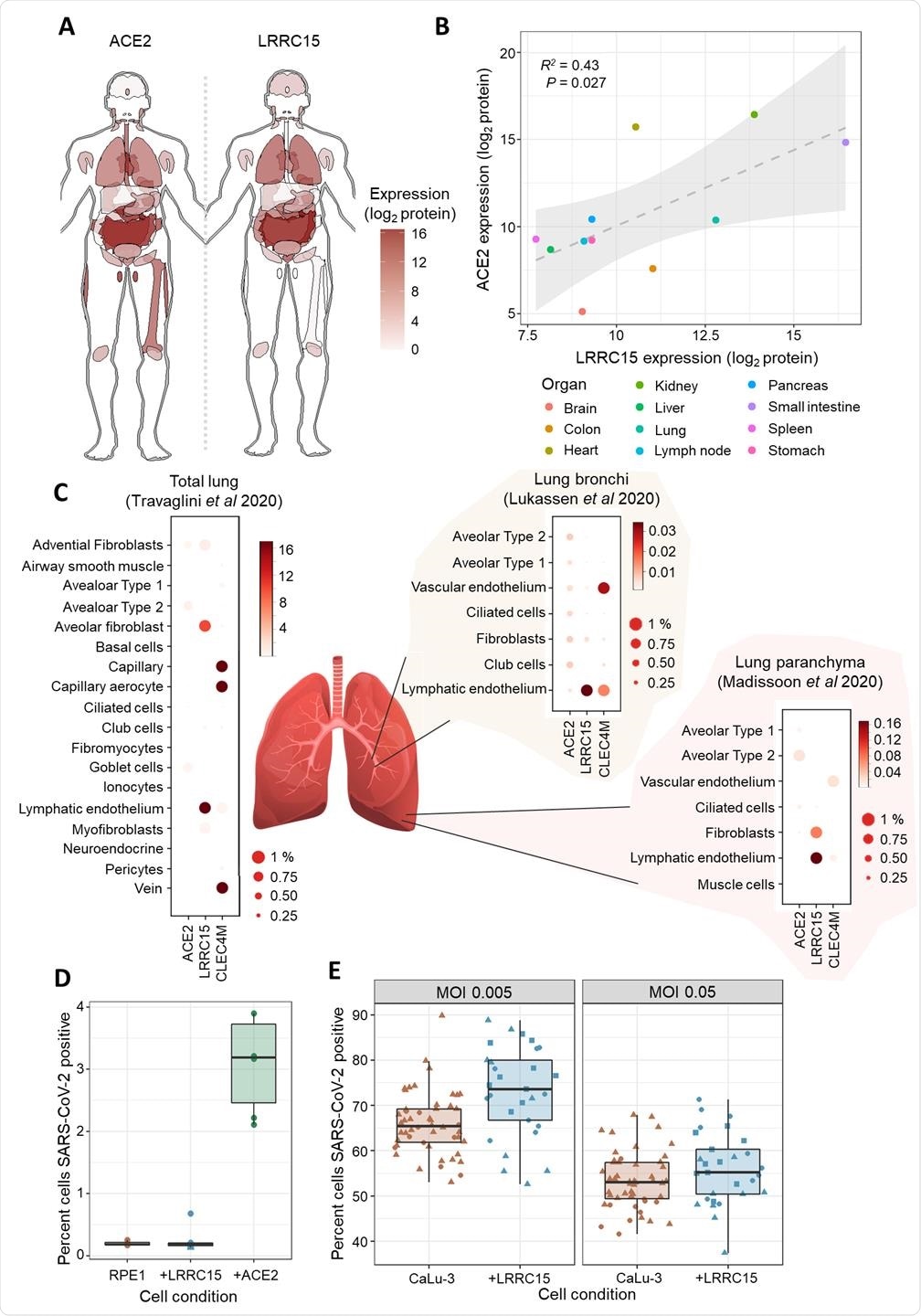
Using proteomics and single-cell transcriptomics, the researchers found that LRRC15 expression was common in human lung vasculature cells and fibroblasts.
While infection assays demonstrated that cell surface expression of LRRC15 alone was not sufficient to permit viral entry, the team found evidence that it could modulate host cells’ susceptibility to SARS-CoV-2 infection.
The spike–LRRC15 interaction “merits further investigation”
Wright and colleagues say that the spike–LRRC15 interaction merits further investigation to determine how SARS-CoV-2 exploits LRRC15 and whether it might account for any distinctive features of COVID-19.
The team also points out that LRRC15 was previously found to be significantly upregulated in the lungs of patients with fatal COVID-19, further emphasizing the value of investigating the role this protein plays in COVID-19 pathology.
Furthermore, several successful therapeutics work by blocking spike’s interactions with host cell receptors, mainly in the form of ACE2-blocking agents. The identification of a shared binding domain between LRRC15 and ACE2 suggests that many of these drugs may also prevent the spike–LRRC15 interaction.
The researchers say that their studies have only just begun to reveal the roles that LRRC15 may play in COVID-19.
“Characterizing the functional and molecular underpinnings of this interaction may provide useful targets for therapeutics and guide our understanding of what makes SARS-CoV-2 so distinctly pathogenic,” they conclude.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Wright G, et al. LRRC15 mediates an accessory interaction with the SARS-CoV-2 spike protein. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.09.25.461776, https://www.biorxiv.org/content/10.1101/2021.09.25.461776v1
Posted in: Medical Research News | Disease/Infection News
Tags: ACE2, Angiotensin, Angiotensin-Converting Enzyme 2, Cell, Coronavirus, Coronavirus Disease COVID-19, CRISPR, Cytometry, Drugs, Enzyme, Flow Cytometry, Gene, Genes, Genome, Glycan, Leucine, Lung Vasculature, Lungs, Mass Spectrometry, Membrane, Microscopy, Pathology, Protein, Protein Expression, Proteomics, Receptor, Research, Respiratory, RNA, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spectrometry, Spike Protein, Syndrome, Therapeutics, Transcription, Transcriptomics, Vasculature, Virus

Written by
Sally Robertson
Sally first developed an interest in medical communications when she took on the role of Journal Development Editor for BioMed Central (BMC), after having graduated with a degree in biomedical science from Greenwich University.
Source: Read Full Article