Genomics is the study of all of a person’s genes, including interactions of those genes with each other and with the person’s environment. Sequencing and analyzing genomic information can help us better diagnose, treat and even prevent disease. However, researchers often have specific questions about the genome. They need access to large and diverse datasets to find the answers.
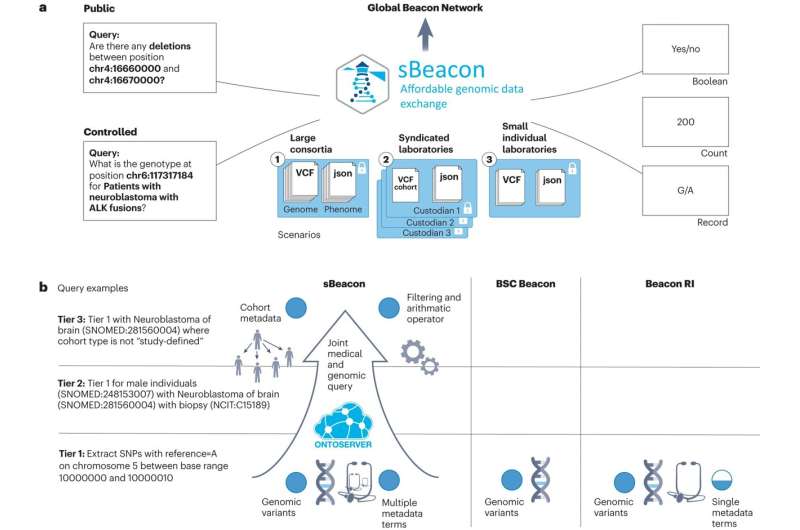
That’s why we’ve created a data-sharing system that enables researchers from around the world to share genomic information. Called sBeacon, the technology enables efficient transfer of genomic data between research institutions.
Now, our software is making genomic data more accessible than ever. It gives smaller and less wealthy countries and institutions equal access to the global network. The sBeacon software has been featured in Nature Biotechnology.
Lighting up genetics research
Enter beacon systems. The technology allows genomic data to be transferred securely between hospitals and research institutions across the globe. Each institution ‘stands up’ a beacon. Researchers or clinicians can activate the beacon to request information. Their request, or ‘query,’ can be answered by a global network of hospitals and research institutions. These institutions light up their own beacon to share specific aspects of their data.
The power of the system increases with the more beacons lit. As of March 2023, only 54 organizations have stood up a beacon.
What if there was a faster, cheaper, and more accessible system? Our researchers decided to make this dream a reality.
Removing beacon system barriers
Dr. Denis Bauer leads our Transformational Bioinformatics group. She is passionate about removing the barriers in genomic data sharing.
“The goal is a future in which every small hospital, health agency, and research facility is able to light up a beacon,” she said.
Her research group worked with the Australian Genomics Health Alliance and Genomics England to develop Serverless Beacon, or “sBeacon.”
“sBeacon can rapidly find and isolate the disease-causing genomic mutations among the three billion letters of the human genome,” Denis said.
Unlike other Beacon systems, sBeacon is cloud-native. This means it does not rely on a central database. The owner of the data retains control over the data. This keeps patients’ information safe and secure.
sBeacon has another unique feature. Institutions are only charged when their beacon is queried. This makes sBeacon 1,800 times faster than other implementations and around 10 times cheaper.
“sBeacon lowers the barrier to entry for smaller or economically disadvantaged organizations to stand up a beacon,” Denis said.
A beacon of hope
Denis hopes that sBeacon will make genomic data sharing more accessible.
“Currently, the number of participating research institutions is small. The data shared within the network is not representative of genetically diverse cohorts. We want to change that.”
Historically, less genomic data has been available from non-Caucasian groups. By enabling more groups to join the beacon network, researchers and clinicians will have access to more diverse data.
“It’s crucial that clinicians and researchers have access to data that is representative of the population so they can more effectively treat all patients who walk through their doors,” Denis said.
Researchers and clinicians can query medical data through sBeacon alongside a patient’s genetic information and adhere to emerging data standards.
“sBeacon offers real-time analysis of the data. With this technology, we can potentially improve the treatment of complex diseases like cancer and cardiovascular disease.”
More information:
Anuradha Wickramarachchi et al, Scalable genomic data exchange and analytics with sBeacon, Nature Biotechnology (2023). DOI: 10.1038/s41587-023-01972-9
Journal information:
Nature Biotechnology
Source: Read Full Article