In a recent study by US researchers (currently available on the medRxiv* preprint server), a specific immunoassay based on the spike glycoprotein’s receptor-binding domain (RBD) of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is not only well suited for the detection of antibody response after infection and vaccination but may also represent a method of choice when conducting seroepidemiological studies.
Following the burden and spread of SARS-CoV-2 infection during the coronavirus disease (COVID-19) pandemic is of utmost importance in small environments, but also larger geographical settings. Basically, all of our current data is derived following the use of two different classes of assays.
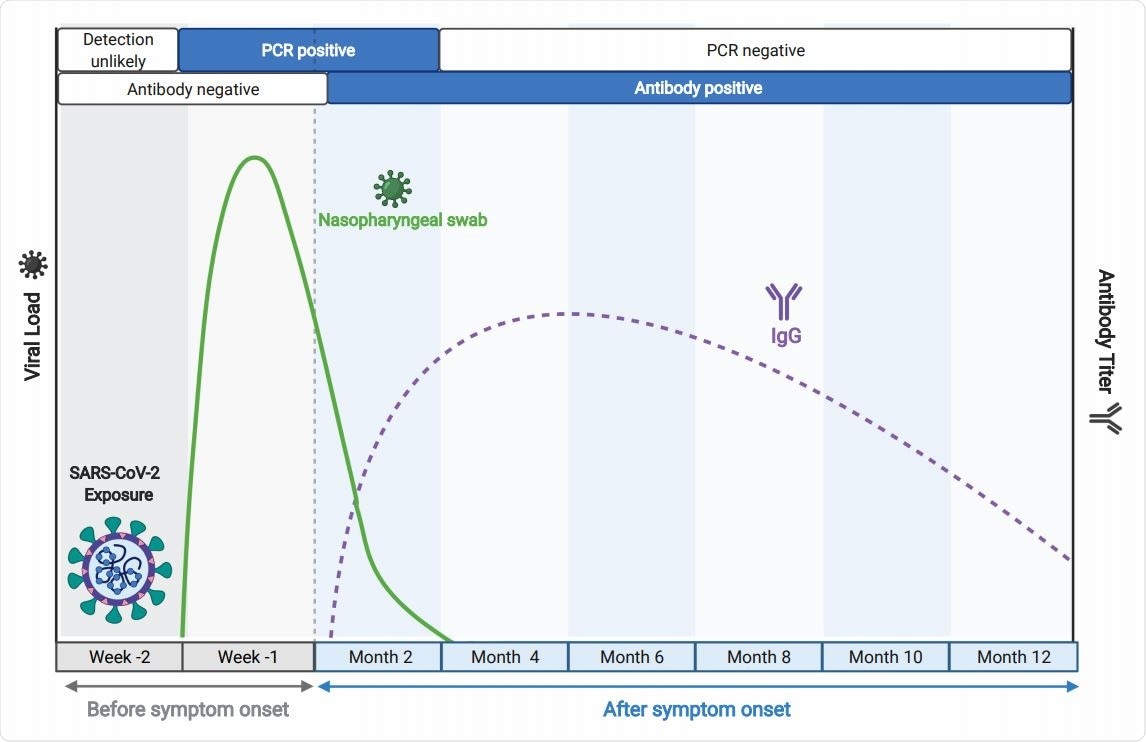
Active disease is diagnosed with the methods that detect the presence of virus in upper respiratory specimens – most notably viral nucleic acid amplification tests (with real-time polymerase chain reaction being the gold standard) and immunodetection of viral antigen.
Conversely, we can also detect the response of our immune systems by detecting virus-specific antibodies, which is done by serological or antibody-based assays. These are precisely the methods that should be used in seroepidemiology, as they can demonstrate prior infections as well.
Antibody-based assays for SARS-CoV-2 are based on two viral antigens: spike glycoprotein that attaches to the host cell receptor (angiotensin-converting enzyme 2 or ACE2) by utilizing RBD in the S1 subunit, and nucleocapsid, which interacts with viral RNA inside the envelope.
Thus far, assays that used nucleocapsid antigen had a preference in a majority of clinical settings, while spike glycoprotein has been chiefly used as antibody capture antigen in research milieus. But is it time for more pervasive clinical applications of spike glycoprotein-based serological assays?
In this new paper, a research group led by Dr. Pratik Datta delineates key traits of their serological enzyme-linked immunosorbent assay (ELISA), which utilizes a novel S1 RBD antigen, as well as its propensity to detect antibodies in minimal amounts of remotely collected peripheral blood.
A comparative study approach
The assays described in this study were performed using a novel gp70-fusion protein form of the S1 RBD antigen. The gp70 domain is characteristic for its chaperone-like qualities, which means it aids in the assembly or disassembly of other macromolecular structures.
Consequently, the researchers compared detection properties of virus-specific antibody responses to SARS-CoV-2 infection of their RBD-based ELISA assay with two specific assays from Roche Elecsys and Abbott Architect that utilize nucleocapsid antigen as the capture reagent.
Furthermore, since different study designs can result in specimen collection of either serum or plasma from peripheral blood, this research group has also conducted a matrix equivalency test to see whether the assay is amenable to different types of liquid specimens (including non-blood bodily fluids).
Characteristics of a versatile assay
In short, albeit the sample size was somewhat limited in this study, the results have shown that this RBD-based ELISA assay has excellent sensitivity and specificity, as well as a favorable profile in comparison compares with commercial tests that are currently widespread in clinical settings.
Moreover, the researchers have shown that microsampling and phlebotomy can be used interchangeably for peripheral blood collection. In contrast, the utilization of serum or plasma from different blood collection tubes had no demonstrable effect on antibody-binding results.
The results also imply that this ELISA protocol can be compatible with various matrices, including serum, plasma acquired using different anticoagulants, and non-blood bodily fluids such as breast milk. All of this makes it very adaptable to a diverse array of study designs.
Improving seroepidemiology research
Overall, the paper neatly demonstrates how this type of ELISA test based on spike glycoprotein RBD can be accurate, adaptable, and highly fitting for a myriad of research and clinical applications.
“Our protocol is performed utilizing robotic sample handling and dilution and automated ELISA,” further explain the authors of this bioRxiv manuscript. “Moreover, it compares favorably with accurate commercial tests that have been widely used in clinical practice to determine exposure to SARS-CoV-2”, they add.
All of this means that its use is remarkably suited for seroepidemiology approaches and other large-scale studies that necessitate parsimonious sample collection outside of healthcare settings. Considering the further spread of SARS-CoV-2, practical and versatile solutions are a welcome addition to our armamentarium.
*Important Notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Datta, P. et al. (2021). Highly versatile antibody binding assay for the detection of SARS-CoV-2 infection. medRxiv. https://doi.org/10.1101/2021.07.09.21260266, https://www.medrxiv.org/content/10.1101/2021.07.09.21260266v1
Posted in: Device / Technology News | Medical Research News | Disease/Infection News
Tags: ACE2, AIDS, Angiotensin, Angiotensin-Converting Enzyme 2, Antibodies, Antibody, Antigen, Assay, Blood, Breast Milk, Cell, Coronavirus, Coronavirus Disease COVID-19, Enzyme, Glycoprotein, Healthcare, Immunoassay, Nucleic Acid, Pandemic, Phlebotomy, Polymerase, Polymerase Chain Reaction, Protein, Receptor, Research, Respiratory, RNA, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Virus

Written by
Dr. Tomislav Meštrović
Dr. Tomislav Meštrović is a medical doctor (MD) with a Ph.D. in biomedical and health sciences, specialist in the field of clinical microbiology, and an Assistant Professor at Croatia's youngest university – University North. In addition to his interest in clinical, research and lecturing activities, his immense passion for medical writing and scientific communication goes back to his student days. He enjoys contributing back to the community. In his spare time, Tomislav is a movie buff and an avid traveler.
Source: Read Full Article