 insights from industryJason Otterstrom PhD and Alexandra Lubin PhDApplication Scientist and Research AssociateIDEA Bio-Medical and UCL Cancer Institute
insights from industryJason Otterstrom PhD and Alexandra Lubin PhDApplication Scientist and Research AssociateIDEA Bio-Medical and UCL Cancer InstituteIn this interview, News-Medical talks to Dr. Alex Lubin and Dr. Jason Otterstrom, about the use of deep learning powered automated microscopy and image analysis for in vivo Zebrafish screening.
Video credit: IDEA Bio-Medical
What is the typical process for imaging zebrafish?
Young zebrafish embryos are taken at around three days post fertilization and placed into a 96-well plate. We use one embryo per well, loaded via standard pipette with a wide bore tip.
The embryos can then be imaged with several types of microscopes and techniques, for example, using brightfield or fluorescence. We use the Hermes WiScan® automated high content screening microscope from IDEA Bio-Medical.
We in the Payne group take four overlapping images along each well at five different z-slice positions. This captures the whole embryo – an important consideration because it is a 3D animal. The most in-focus slice is chosen of the brightfield images, and we perform maximum intensity projections on the fluorescent images. We use Hermes software to batch process our datasets.
The resulting montages provide two images for each embryo – one in brightfield and one in fluorescence. We use a transgenic green fluorescent protein (GFP) which has the gene CD41 tagged with green fluorescence to label hematopoietic stem cells. We are interested in visualizing and measuring the number of GFP-cells located specifically in the tail and study how different compounds affect them.
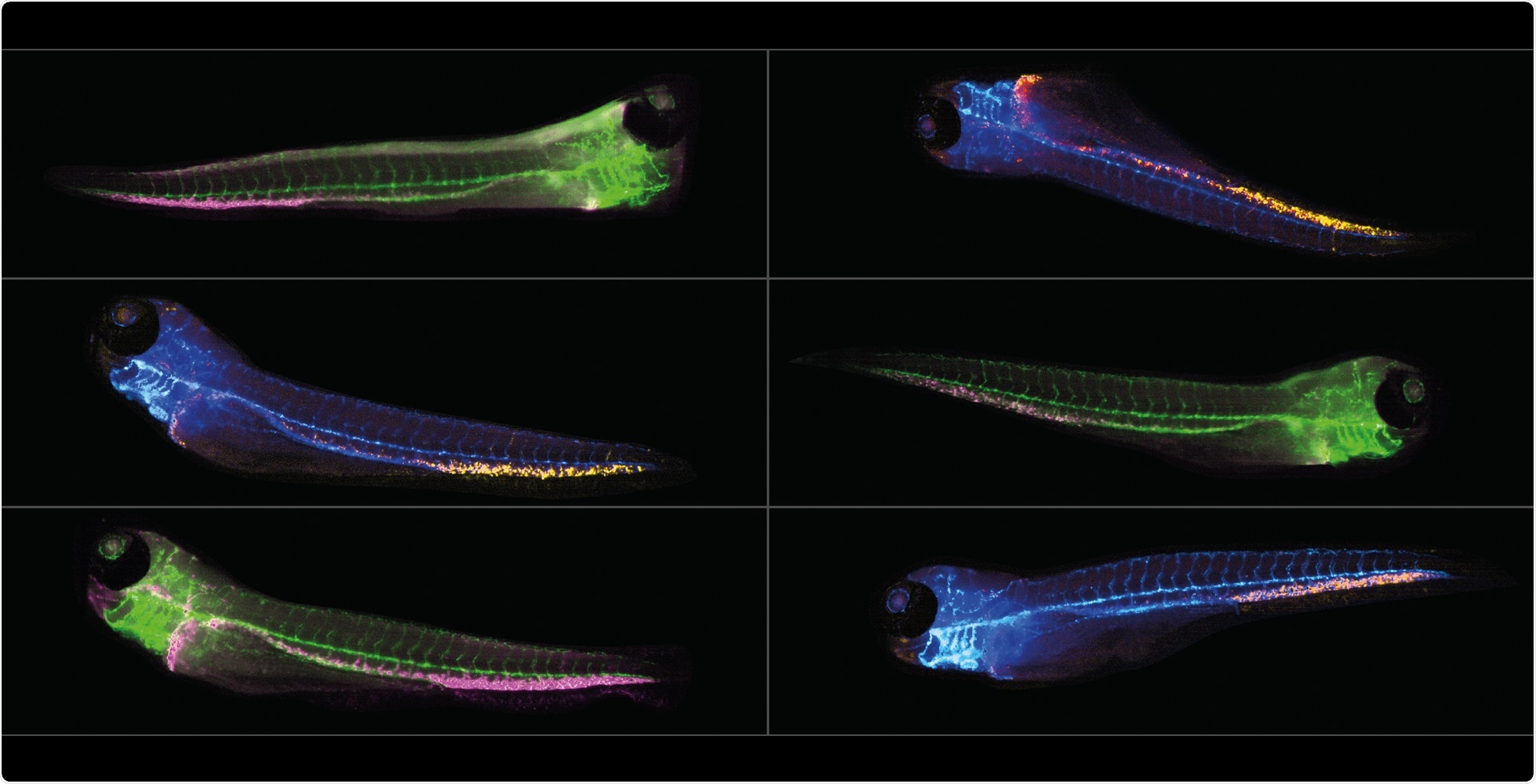
Image credit: IDEA Bio-Medical
What is the biggest difficulty you encounter with zebrafish?
For us, imaging of large numbers of zebrafish is relatively straightforward. We encountered major bottlenecks when looking to automate analysis of the images. Ultimately, we overcame this challenge by collaborating with IDEA Bio-Medical to help develop an automated analysis application within their WiSoft® Athena software.
Regarding the difficulties in analysis, the first is that these fish are living and 3D, so they are unlikely to be positioned perfectly or uniformly in the wells. In a large-scale automated drug screen, it is impractical to go through the images manually to exclude those fish that are improperly positioned.
Another key analysis challenge is the ability to examine one specific anatomical region of the zebrafish embryo, while ignoring others. For example, when looking at cells in the tail, standard fluorescence analysis using thresholding picks up bright spots elsewhere in the fish, either from cells located in the blood or autofluorescence.
It is common to manually select the region of interest, but this makes it impossible to automate the workflow and reduces the accessible drug library size.
Another method involves adapting software developed for cell-based screening, particularly software that allows users to look for fluorescent structures within other structures. However, we still found that this was picking up fluorescence in the other areas that we were not interested in. None of these methods were wholly suitable.
Could you elaborate on how the Hermes WiScan microscope and the WiSoft Athena software support your study of zebrafish embryos?
The WiScan® Hermes is a fully automated microscope for high content screening. If any of your readers are unfamiliar with high content screening, it is the automated and unbiased acquisition and analysis of microscopy images.
This methodology is commonly used in drug discovery to identify and confirm biologically active compounds that cause a biological effect which is visualized within the microscopy images.
The Hermes platform consists of the automated microscope and the accompanying WiSoft Athena image analysis software. Together, these enable the imaging and analysis of a broad range of biological samples with minimal user intervention.
The Hermes is an inverted wide-field microscope. Its most unique aspect is that the objective is moving during a sample scan, leaving the sample remains largely stationary.
This means that a plate or other sample type is not jostled about or shaken, as is commonly the case when the microscope stage is used for sample scanning. Moving the objective and not the sample ensures gentler handling during the imaging of delicate samples. The objective can move in all three directions – X, Y and Z – so z-stack acquisition is supported.
Since the objective is relatively small and light, it also enables rapid scanning of a broad range of samples. The Hermes can scan a 96-well plate in just under 2 minutes and a 384-well plate in about 5 minutes and 30 seconds. Such reference scans are set to image in four colors with one field-of-view & z-slice per well with 50 milliseconds exposure per color.
The system also includes a laser-based auto focus that is compatible with round-bottom plates. Round-bottom plates are commonly used with 3D cell cultures, but these have also been used as a novel method to orient zebrafish and facilitate imaging of their head/brain region.
The Hermes supports objectives with magnifications from 2X to 60X, and we utilize only the highest quality Olympus objectives. Our standard system is compatible with air objectives, and we offer upgrades to permit the use of oil or water immersion objectives.
Designed specifically for zebrafish researchers, the Hermes Zebrafish Partner contains all the elements necessary for automated full-plate scanning and image analysis of zebrafish larvae. It has two fluorescence colors plus brightfield, a single objective of choice and an attractive price point that fits into research budgets.
How are zebrafish images analyzed in the Athena software?
The zebrafish-specific platform was inspired by the needs of researchers who use this biological model system. Their bottlenecks and challenges prompted us at IDEA Bio-Medical to develop a new solution to fully automated high content imaging and analysis of zebrafish embryos. With this platform, we aim to facilitate drug, genetic or toxicological screening studies that use fish and thereby increase the accessible throughput.
The Athena’s underlying deep learning AI allows parameter-free analysis of zebrafish morphology and anatomy in brightfield images. The AI not only outlines the fish contour – but it also identifies the eye, the otic vesicle, the yolk sac, the swim bladder, the heart, the spine or notochord, the tail fin and three body regions: the head, the trunk and the tail.
All these structures are identified in simple brightfield images without the need for user-defined parameters. And for each structure, the software quantifies morphology in terms of the area, the perimeter and the shape. Each object that is automatically identified in the fish can also be manually segmented or edited using the software’s Manual Analysis tools.
Brightfield image structure and information is combined with fluorescence data to extract rich and anatomically relevant information. The software quantifies the total fluorescence intensity and/or count of the number of fluorescent objects identified within each structure.
Once analyzed, fish can be classified after processing to identify populations or groups that are of interest or select desired embryo orientations. Lastly, the imaging and analysis can be used with time-lapse and/or z-stack images.
Overall, the biggest advantage is that it only takes about an hour to fully process a 96-well plate, including imaging and analysis. In detail, we need approximately 5 minutes to prepare the plate, around 15 minutes for imaging, 10-20 minutes to process the images and another 10-20 minutes to run the Athena software to get the data out. This throughput makes it feasible to screen larger compound libraries and moves the workflow bottleneck to other sample handling and processing steps.
Could you tell our readers more about the use of the Hermes platform in practice?
One project where we apply the zebrafish model, and the workflow we have described, is to study two types of blood cancer. Myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) are most common with aging, occurring due to mutations in the hematopoietic stem and progenitor cells.
In our lab, we are particularly interested in a phenomenon called CHIP (clonal hematopoiesis of indeterminate potential) – an age-related phenomenon where, as you develop mutations naturally as you age, you develop a ‘driver mutation’ that gives those cells a clonal advantage.
If cells with this mutation develop, the cell population suddenly starts to expand significantly. MDS and AML can then develop if this clonal population gain further mutations.
There are several driver mutations in genes, including DNMT3A and ASXL1. Our goal is to perform a drug screen that targets mutations in these genes and clonal populations.
This could potentially be used as a therapeutic for MDS or AML, particularly as the therapeutics for these diseases are often very invasive and not suitable for people who are older with slightly milder disease. It could also potentially be used in a pre-malignant state before disease develops when the initial clonal expansion is occurring.
The hematopoietic system in zebrafish is remarkably similar to that of humans. They have all the same types of blood cells as humans, but they develop much faster.
Zebrafish go into ‘definitive hematopoiesis’ from about 48 hours, which means we can look at hematopoiesis in these samples whilst they are relatively young, transparent embryos. For this project, we opted to evaluate this after three days.
The gene we are focusing on initially is DNMT3A, which encodes for a DNA methyltransferase. This is the most commonly mutated driver gene in MDS and AML.
There is a particular hotspot called R882 that is commonly mutated. Zebrafish have two copies of DNMT3A – DNMT3AA and AB. There is a very good correlation between these two orthologues and the human gene, with an identical amino acid sequence present at the R882 hotspot.
We have used CRISPR in one-cell stage embryos to generate some knock-out lines for DNMT3AA and AB around this R882 hotspot. We have generated a stable knock-out line for both orthologues.
On day 3, the HSCs (hematopoietic stem cells) that we are interested in are located in the caudal hematopoietic tissue (CHT) at the very base of the tail of the zebrafish. We use a transgenic fluorescent line called CD41:GFP to visualize the CD41-expressing stem cells and investigate their numbers.
Our goal, in terms of drug screening, is to take wild-type embryos and the DNMT3A mutants we have made, treat these with a variety of drugs from a drug library, and look for compounds that cause a depletion in the number of mutant cells but leave the wild-types unaffected.
Essentially, we are looking for synthetic compounds that only target the mutant cells.
To investigate this, we image, analyze and quantify the embryos with the Hermes WiScan using the approach outlined earlier. We ideally want the zebrafish to be aligned on their side, so we use zebrafish alignment plates from a company called Hashimoto.
Our overall goal is to count the stem cells of interest. We must count fluorescent spots in the fish, but only keep those present in the tail and from embryos properly oriented on their side. Athena can achieve all these requirements in a fully automated fashion.
Once you had the stem cell counts from the study, how did you analyze and confirm the accuracy of these?
The first thing we wanted to do was see how well these results correlated with manual counting. We found that this correlated well with a Pearson Correlation Coefficient of 0.84.
We also wanted to look at different ages; for example, the stem cells start to migrate to the CHT region at two days. The population there starts to increase over time – a pattern which we can see clearly and track with the Athena software.
It was also important to evaluate things that we already knew would reduce the number of stem cells to confirm biological relevance. X-ray radiation is known to destroy stem cells, so we treated zebrafish embryos with X-ray radiation and imaged them at three days.
We confirmed that the software extracted the expected large drop in the stem cell numbers with mid-range radiation dose, and detected a smaller drop with higher dose.
We also looked at a more subtle stem cell phenotype type using another mutant that we had in our lab with a known phenotype; an RPS14 mutant – a ribosomal protein that is also linked to MDS. In a knock-out line, we detected no phenotypic difference between the heterozygous and wild-type embryos without stress.
We discovered that following application of phenylhydrazine (a haemolytic stress), which causes anemia, only the wild-type embryos recover through an increase in stem cells to produce more red blood cells. We did not observe recovery in the mutants[i].
We observed and investigated all these processes using Hermes and Athena together, highlighting how useful this platform is in drug screening applications such as ours.
What about drug screening, have you tried screening your fish lines in this context?
To perform the drug screening itself, we start with the DNMT3AA wild-type and AB wild-type siblings to get embryos that are known to be wild-type. We then took the homozygous DNMT3AA and crossed these to the homozygous DNMT3AB, resulting in double heterozygous embryos for both 3AA and 3AB. This combination allowed us to drug treat these to look for compounds that cause a reduction of stem cells in the double heterozygous animals, without affecting the wild type embryos.
At 24 hours, we treated the embryos with PTU to prevent pigment format, dechorionated them and then placed these into 12-well plates where we treated them with drugs for 48 hours. Drugs were defined by the Tocris library initially, which is a library of 1,120 biologically active compounds. The embryos are loaded into alignment plates for imaging on day 3.
Running the imaging in the Athena allows the acquisition of HSC counts. So far, we have tested around 400 compounds. There have been some initial hits, but none of these have been retested at this stage.
What sort of other applications or analyses can be completed using the Hermes and Athena platform?
Whilst we were using the Athena, we began to realize how flexible and easy to use it was – even for researchers without a background in microscopy or image analysis.
We began to explore its suitability for other applications and have just had a paper accepted for publication on this subject[ii].
We investigated the platform’s potential for working with double transgenics since the Hermes is capable of imaging many different colors and the Athena can analyze these images.
We discovered that it was possible to use double transgenics to look at different types of blood cell in the same animal and tested them using irradiation.
We also looked at the potential for analyzing acridine orange – a fluorescent apoptosis marker used in toxicity screening. There, we confirmed the quantification accuracy using irradiation to induce and measure cell death.
Another quite common fluorescence assay is known as a hair cell assay because it highlights fluorescent hair cell markers. These assays are used to look for ototoxicity, and it was possible to investigate this fluorescence using the Hermes and Athena platform.
When looking to confirm the accuracy of these analyses, we took pre-published data from 2008 and attempted to recreate the results, aiming to see if it was possible to achieve the same results with the Hermes as the study had via manual image analysis.
We adjusted the parameters to look at the larger fluorescent granules, and then we looked at the total intensity of them and the granule area, confirming that both parameters could replicate the results found in the publication.
Another interesting application we evaluated was the platform’s potential to measure angiogenesis and inhibition of angiogenesis. This is of interest for the treatment of solid tumors, and there are several anti-angiogenic compounds currently used in cancer treatment.
We also used pre-published data to confirm accuracy here, using data from a different type of automated screen performed in 2007. The study focused on two compounds that increase in anti-angiogenic properties with increasing concentration.
We were able to set Athena to measure the blood vessels and looked at the total mCherry area in the fish as defined by those blood vessels. We were able to confirm that the reduction caused by these compounds matched the results from the published data.
Finally, I should point out that the Athena software can accept images from third-party microscopes. We are currently preparing a new product that is a standalone version of our analysis software. This software is designed to work specifically with images that are not from the Hermes.
About the interviewees
Dr. Otterstrom is an application scientist for IDEA Bio-Medical. He has a diverse background in biophysics including microscopy, optical design, image analysis and sample labeling. His expertise is adapting biological assays to benefit from utilization of automated microscopy methods.

Dr. Lubin is a post-doc at the UCL Cancer Institute where she uses zebrafish to study myelodysplastic syndrome (MDS) and acute myeloid leukaemia (AML), aiming to develop novel therapeutic treatments. Previously, she obtained her PhD in Chemical Biology from Imperial College London after studying Chemistry at the University of Cambridge.

About IDEA Bio-Medical Ltd.
IDEA Bio-Medical is founded in 2007 through a partnership between Weizmann Institute of Science and IDEA Machine Development.
The company specializes in automated imaging systems and image analysis software, offering a broad range of biological applications based on the company’s unique algorithms library.
The WiScan Hermes system incorporates the most advanced technologies for fast plate scanning to generate sharp, high-contrast images. The easy-to-use WiSoft® Athena software maintains high sample throughput with a fast-analysis algorithms library. Specifically for zebrafish research, the AI-powered zebrafish analysis application quickly and easily quantifies anatomically relevant metrics from large, montaged images. The Hermes platform enables high-impact science, illustrated through the more its than 100 citations from researchers around the globe.
[i] Peña, O.A.; Lubin, A.; Hockings, C.; et al. TLR7 ligation augments hematopoiesis in Rps14 (uS11) deficiency via paradoxical suppression of inflammatory signaling. Blood Adv. 2021, 5(20): 4112-4124. DOI: https://doi.org/10.1182/bloodadvances.2020003055
[ii] Lubin, A.; Otterstrom, J.; Hoade, Y.; et al. A versatile, automated and high-throughput drug screening platform for zebrafish embryos. Biol Open. 2021, 10(9): bio058513. doi: https://doi.org/10.1242/bio.058513
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
IDEA Bio-Medical Ltd.
